This plasmid map creation guide covers key concepts such as restriction enzymes, gel electrophoresis, Southern blotting, plasmid cloning, bacterial transformation, and colony hybridization. You’ll learn how to prepare DNA samples, transfer them to a solid support, and use labeled probes to detect specific sequences. The guide also provides an overview of plasmids and their role, as well as techniques for inserting DNA fragments into plasmids. Additionally, it covers the process of bacterial transformation and how to screen bacterial colonies for the presence of specific DNA sequences. By integrating these concepts, you’ll be able to determine the physical arrangement of a plasmid, creating a detailed plasmid map.
Understanding the DNA Toolkit: A Journey into Molecular Biology
In the vast world of molecular biology, there lies a hidden toolkit that empowers scientists to investigate and manipulate DNA, the blueprint of life. This toolkit holds techniques that allow us to unravel the secrets of DNA’s intricate sequences, paving the way for breakthroughs in medicine, biotechnology, and our understanding of the fundamental building blocks of life.
Essential Tools: Restriction Enzymes and Gel Electrophoresis
Among the most indispensable tools in this molecular toolbox are restriction enzymes, molecular scissors that recognize and cut DNA at specific sequences. This precise cutting ability allows researchers to isolate and analyze DNA fragments of interest. Hand in hand with restriction enzymes goes gel electrophoresis, a technique that separates DNA fragments based on their size and charge. This process enables scientists to visualize these DNA fragments, yielding valuable information about their length and composition.
Unraveling DNA’s Secrets: Southern Blotting
The next step in our molecular journey is Southern blotting, a technique that empowers us to identify specific DNA sequences. By immobilizing DNA fragments on a solid support and using labeled DNA probes, researchers can detect the presence of target sequences. This powerful method has revolutionized genetic testing, allowing for the diagnosis of inherited disorders and the identification of disease-causing mutations.
Plasmid Cloning: Nature’s Molecular Clipboard
A key component in the molecular toolkit is plasmid cloning. Plasmids are small, circular DNA molecules that can replicate independently within bacteria. Scientists have harnessed the natural ability of plasmids to carry foreign DNA, making them indispensable tools for gene manipulation and protein production. Using restriction enzymes and ligase, researchers can insert DNA fragments into plasmids, creating recombinant DNA molecules that encode proteins of interest.
Southern Blotting: Unraveling DNA’s Secrets
Delving into DNA Analysis
Genetics, the study of inheritance, has revolutionized our understanding of life. One essential technique in this field is DNA analysis, which allows us to study the structure and sequence of DNA, the blueprint of life. Southern blotting is a fundamental technique in DNA analysis, enabling us to uncover specific DNA sequences within a vast genome.
Preparing the DNA Sample
The journey begins with a DNA sample, usually extracted from cells. To make Southern blotting possible, the DNA must be broken down into smaller fragments using enzymes called restriction enzymes. These enzymes recognize and cleave DNA at specific sequences, generating a collection of fragments with varying lengths.
Transferring the Fragments
Once the DNA is fragmented, it’s time to transfer it to a solid support. This is achieved by gel electrophoresis, a technique that separates DNA fragments based on their size. The fragments are loaded onto a gel, and an electric current is applied, causing the fragments to migrate through the gel. Smaller fragments move faster, while larger ones lag behind. This separation creates a distinctive pattern of DNA bands visible under ultraviolet light.
Detecting Specific Sequences
The next step is to identify specific DNA sequences within the mix of fragments. This is where labeled probes come into play. Probes are complementary DNA or RNA sequences that are designed to bind to specific target sequences within the DNA sample. The probes are labeled with radioactive or fluorescent markers, making them visible when they hybridize with their target sequences.
Visualizing the Results
The final step involves transferring the separated DNA fragments from the gel to a solid support, typically a nylon membrane. The membrane is then incubated with the labeled probe, allowing the probe to bind to its complementary sequences. The membrane is then washed to remove unbound probes, leaving only the bound probes visible when exposed to X-ray film or a scanner. The resulting pattern of labeled bands reveals the location of specific DNA sequences within the original sample.
Southern blotting is a powerful technique that provides valuable insights into the structure and makeup of DNA. From disease diagnosis to forensic investigations, this technique continues to play a vital role in the field of genetics. By understanding the intricacies of Southern blotting, we can further our exploration into the mysteries of life and unlock the potential of genetic information.
**Plasmid Cloning: The Art of Inserting DNA into Bacterial Hosts**
In the realm of molecular biology, we often find ourselves manipulating DNA to gain insights into its structure and function. One essential technique in this pursuit is plasmid cloning, a process that allows us to insert specific DNA fragments into bacterial hosts, creating “recombinant plasmids.”
Plasmids: Nature’s Tiny Carriers
Plasmids are small, circular pieces of DNA found in bacteria. They are “extrachromosomal,” meaning they exist outside the cell’s main chromosome. Plasmids play a crucial role in the genetic makeup of bacteria, carrying genes that confer specific traits or confer resistance to antibiotics.
Inserting DNA: The Precision of Restriction Enzymes
To insert DNA into a plasmid, we utilize restriction enzymes, molecular scissors that recognize and cut DNA at specific sequences. These enzymes are named after the bacteria from which they were isolated, such as EcoRI and BamHI. By carefully selecting restriction enzymes, we can create “sticky ends” on the DNA fragments and complementary ends on the plasmid.
Ligase: The Master Welder
Once the DNA and plasmid have compatible sticky ends, we introduce the enzyme ligase. Ligase acts as the molecular welder, joining the DNA fragments and plasmid together to form a recombinant plasmid. This process requires careful temperature control to ensure optimal ligase activity.
Bacterial Transformation: A New Home for Recombinant Plasmids
The next step in plasmid cloning is introducing the recombinant plasmids into Escherichia coli bacteria. This process, known as bacterial transformation, involves making the bacterial cells competent, allowing them to take up foreign DNA. Various techniques, such as electroporation or chemical transformation, can be employed for efficient transformation.
Screening and Selection: Finding the Right Bacteria
After transformation, we must screen and select bacterial colonies that successfully incorporated the recombinant plasmid. This is typically done using antibiotics, exploiting the resistance genes carried by the plasmid. By growing the transformed bacteria on growth media containing the appropriate antibiotic, only bacteria containing the plasmid will survive and form colonies.
Colony Hybridization: Confirming DNA Presence
To confirm the presence of the specific DNA fragment in the recombinant plasmids, we perform colony hybridization. This technique involves transferring bacterial colonies onto a filter membrane and then probing the membrane with a labeled DNA fragment complementary to the target sequence. The presence of a colored precipitate indicates successful hybridization, confirming the presence of the desired DNA fragment in the plasmid.
Bacterial Transformation: The Magic of Recombinant DNA
In the realm of genetic engineering, bacteria stand as invaluable partners, playing a crucial role in the transformation of recombinant DNA. The ability to insert foreign DNA into bacteria, known as bacterial transformation, has revolutionized the field of molecular biology.
Meet the Players:
The key component of this process is a recombinant plasmid, a circular DNA molecule that carries the desired genetic material. These plasmids serve as vehicles, delivering the foreign DNA into the bacteria.
The Transformation Process:
The transformation process begins with the preparation of competent bacteria, which are cells that have been made receptive to foreign DNA uptake. Recombinant plasmids are introduced into these bacteria using various techniques, such as electroporation or chemical transformation. During electroporation, a brief electrical pulse momentarily creates pores in the bacterial cell membrane, allowing the plasmids to enter. In chemical transformation, a solution of calcium chloride is used to facilitate plasmid entry.
Antibiotic Resistance: A Guiding Light:
To select for bacteria that have successfully taken up the recombinant plasmid, antibiotics come into play. These drugs kill bacteria that do not carry the plasmid. Plasmids commonly contain antibiotic resistance genes, which protect bacteria from specific antibiotics. By including an antibiotic in the growth medium, only bacteria carrying the recombinant plasmid will survive, creating a selective environment.
The Legacy of Transformation:
Bacterial transformation has played an indispensable role in the advancement of genetic engineering. It has enabled researchers to create genetically modified organisms (GMOs) with altered traits, ranging from improved crop yields to the production of medical therapies. Additionally, bacterial transformation serves as a cornerstone for techniques such as gene cloning, DNA sequencing, and genetic screening.
Bacterial transformation stands as a testament to the ingenuity of scientists in harnessing the power of nature to manipulate DNA. Through this process, we have gained an unparalleled understanding of the genetic code and opened up countless possibilities for innovation in medicine, agriculture, and beyond.
Colony Hybridization: Uncovering the Secrets of Bacterial DNA
In the realm of DNA analysis, colony hybridization stands as a powerful technique for identifying bacterial colonies harboring specific DNA sequences. It’s like a detective searching for clues, but instead of footprints or fingerprints, the evidence lies within the genetic code of bacteria.
Colony hybridization begins with the preparation of a filter paper resembling a miniature crime scene. Bacterial colonies, each containing millions of cells, are carefully transferred onto this filter. These colonies become the suspects in our investigation.
The next step involves DNA probes, specially designed molecular tools that act as precision-guided missiles. These probes carry labeled DNA sequences complementary to the target DNA we’re searching for. When they encounter a matching sequence on the filter paper, they bind to it like detectives finding their mark.
The detectives aren’t finished yet! To visualize the suspects, they expose the filter paper to X-ray film or a similar detection system. The bound probes emit signals that create dark spots on the film, revealing the location of bacterial colonies containing the target DNA. It’s like a molecular treasure hunt, where the darkest spots indicate the most suspects with the desired genetic code.
Colony hybridization is a versatile technique used in various fields like:
- Genome mapping: Identifying the location of specific genes on bacterial chromosomes
- Mutation screening: Detecting changes in genetic sequences
- Pathogen identification: Identifying disease-causing bacteria
By uncovering the secrets of bacterial DNA, colony hybridization aids scientists in advancing our understanding of microbiology, genetics, and the diagnosis and treatment of infectious diseases.
So, next time you hear about colony hybridization, remember this thrilling detective story, where the filter paper becomes the crime scene, and the DNA probes are the detectives searching for the genetic evidence that will solve the mystery.
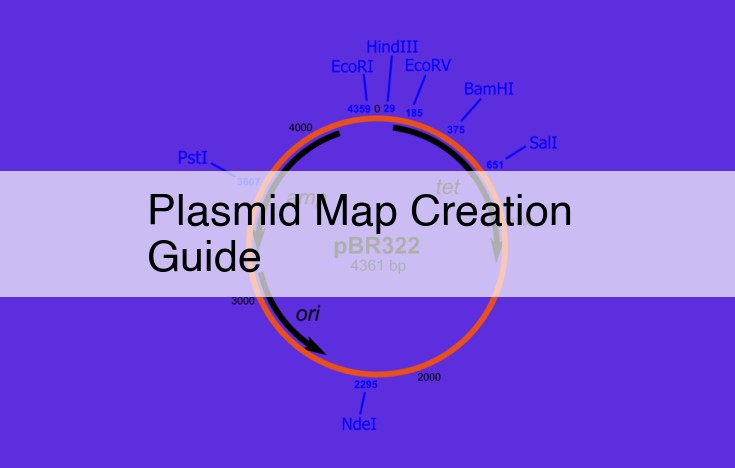
Creating a Plasmid Map: Unveiling the Architectural Blueprint of DNA
In the realm of molecular biology, creating a plasmid map is akin to deciphering a DNA blueprint, revealing the intricate arrangement of genetic material within a plasmid. A plasmid, a small circular DNA molecule found in bacteria, serves as a valuable tool for cloning and manipulating genes, making plasmid mapping a crucial step in understanding and utilizing their potential.
To embark on this mapping endeavor, you’ll need to integrate knowledge gained from previous steps, such as restriction site mapping and orientation determination. Restriction enzymes, molecular scissors, meticulously cleave DNA at specific recognition sequences, generating fragments of varying lengths. By digesting a plasmid with multiple restriction enzymes and analyzing the resulting fragment sizes, scientists can deduce the location and order of restriction sites within the plasmid.
Armed with this information, restriction site mapping provides a linear snapshot of the plasmid’s DNA sequence. However, to truly grasp its architecture, orientation determination becomes indispensable. Ligation, a process that seamlessly connects DNA fragments using an enzyme called DNA ligase, allows researchers to insert specific DNA sequences into a plasmid. These inserted sequences, often carrying antibiotic resistance genes, serve as selectable markers, enabling scientists to identify and isolate bacteria that have successfully taken up the recombinant plasmid.
By subjecting these transformed bacteria to colony hybridization, a technique that employs labeled DNA probes to detect specific DNA sequences, researchers can identify colonies that contain the desired plasmid. This screening process ensures that only colonies harboring the correct plasmid are selected for further analysis.
Finally, integrating all these concepts culminates in the creation of a plasmid map. This map, a comprehensive graphical representation, precisely depicts the physical arrangement of the plasmid, including the locations of restriction sites, selectable markers, and any other relevant genetic features. Armed with this blueprint, scientists can design and execute experiments to manipulate genes within the plasmid, paving the way for groundbreaking discoveries in genetic engineering and biotechnology.